Salmonella enterica
History
The earliest classification of Salmonella was based on serotypes, and according to the modern classification system, the early 99% of the taxon was eventually merged into S.enterica. S.enterica contains six subspecies: enterica, salamae, arizonae, diarizonae, houtenae, and indica. Among them, S. enterica subsp. enterica contains the most known serotypes. Including S. Typhimurium, Salmonella paratyphi is finally classified as the same name of S. enterica subsp. enterica.
Species characteristics
Gram-negative Enterobacter, containing weekly flagella, facultative anaerobic. Adapted to the intestinal growth of animals, the optimal growth temperature is 37 degrees Celsius. Some serotype strains can be adapted to multiple hosts, while other serotype strains may be specific to a particular host. The S. Typhi and S. Paratyphi hosts are generally human. Salmonella can survive for long periods of time in vitro, such as in food processing environments, but rarely through water.
Pathogenicity
The infection of Salmonella is mainly transmitted through the feces-mouth, which causes disease at the end of the small intestine and the upper end of the large intestine after passing through the stomach acid. Non-typhoid Salmonella invades the gastrointestinal tract and causes inflammatory diarrhea. The invasive site is characterized by neutral infiltration, while the inflammatory response caused by Salmonella typhimurium invasion is not obvious, few neutrophils accumulate, and Salmonella typhimurium is expressed. The toxin reduces the host's inflammatory response and invades the endothelium system.
Genome
Until (July 2020 ),17,352 Salmonella enterica genomes have been published on NCBI,the sequences files are fromGenBank。
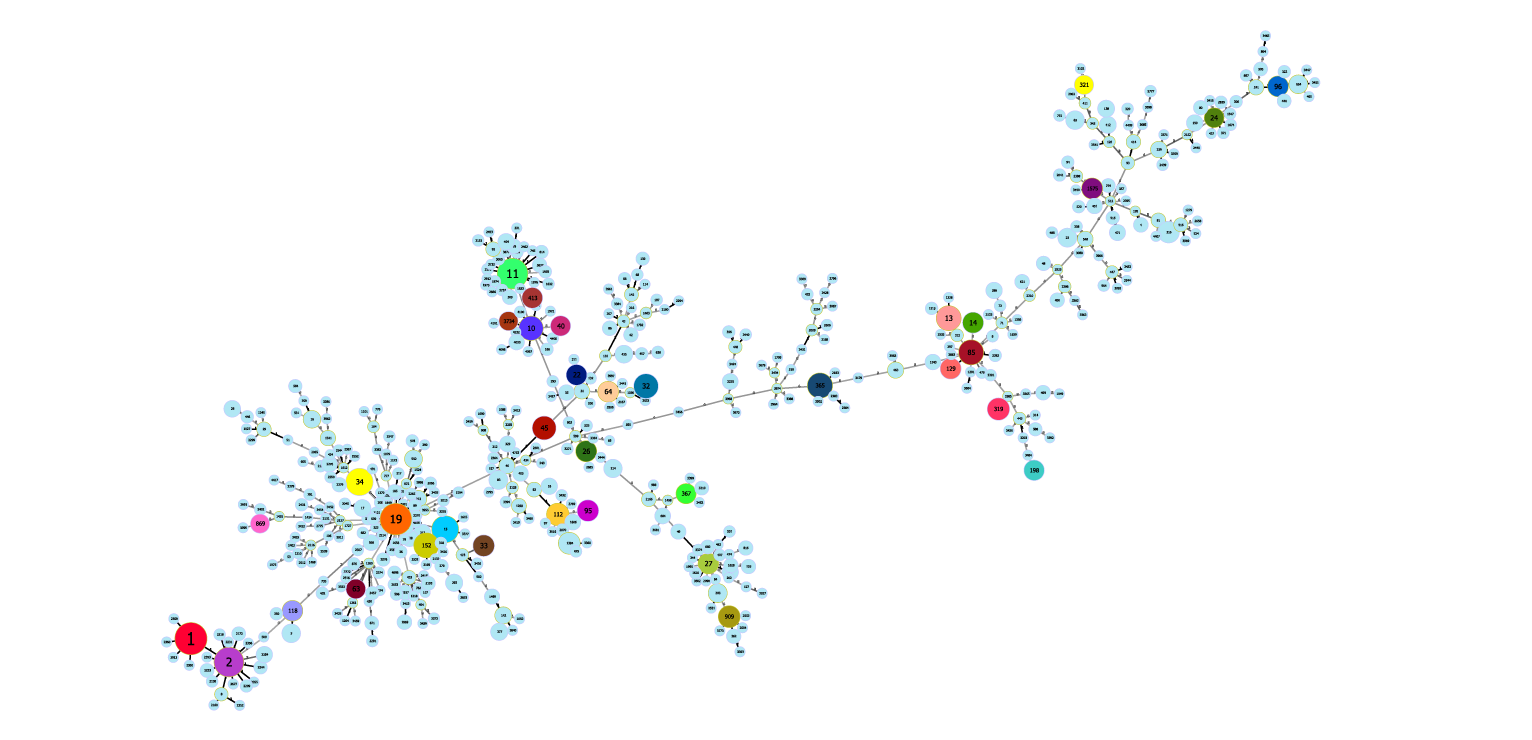
ST types published on PubMLST: http://enterobase.warwick.ac.uk/species/senterica/allele_st_search

Sequence type