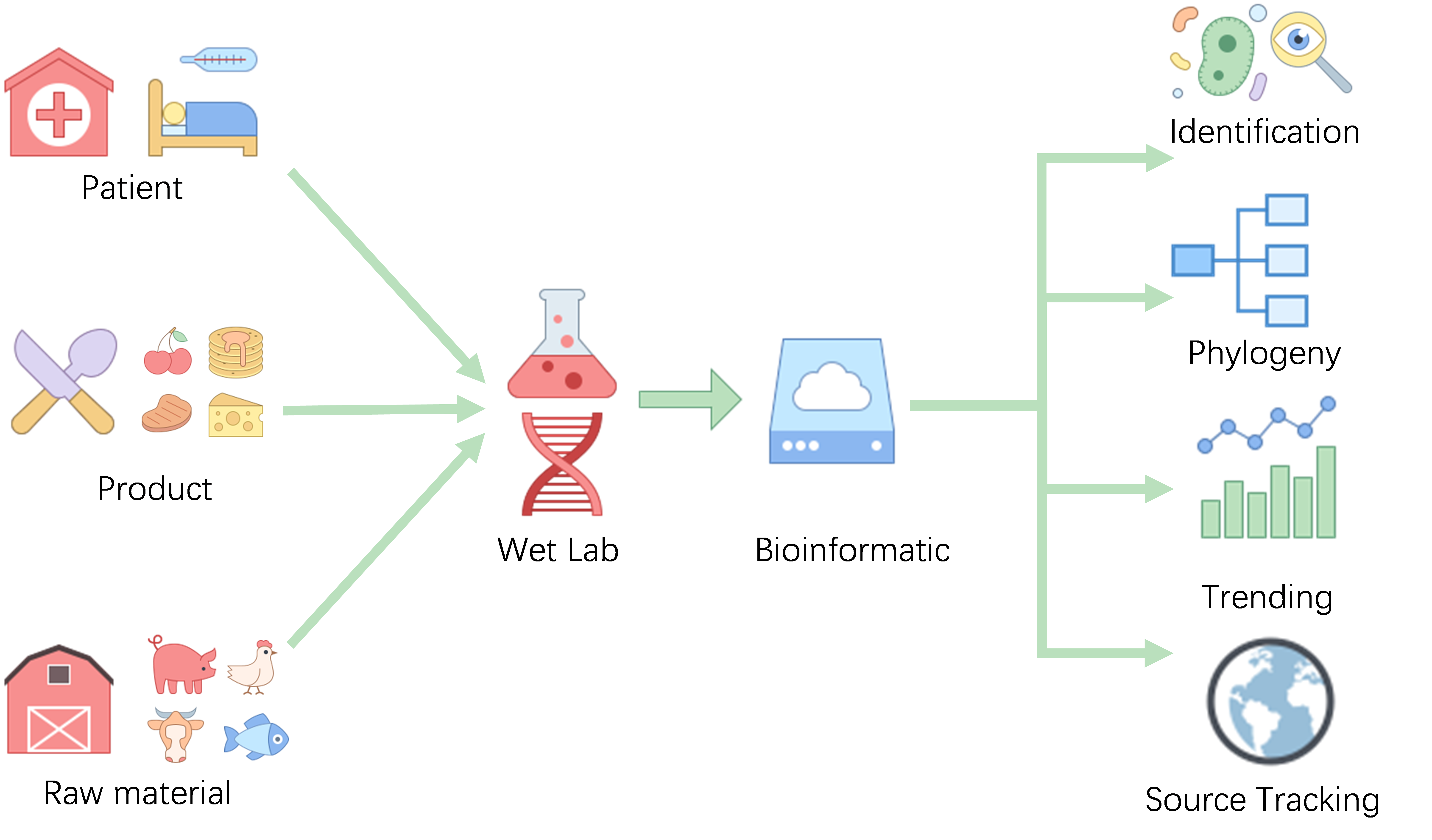
Whole genome data provides more detailed and precise information for pathogen identification and strain typing, playing increasingly important role in food safety or clinical purpose. Available microbial genomic data is growing rapidly, but there are some challenges before application including errors in species label or contamination in the public available genome data, absence of the species context for antibiotic or virulence factor analysis of unknown genome and a typing scheme for pathogen tracking.
PBGdango provide a curated type species reference genome database for accurate bacterial identification and phylogenetic analysis, we corrected genomes of six foodborne pathogens-Vibrio parahaemolyticus, Listeria monocytogenes, Salmonella enterica, Staphylococcus aureus, Bacillus cereus, Clostridioides difficile currently . PBGdango aims to help to tracking the pathogen from raw materials or environment to final product and to the patient. The data page shows the genomic information of the six pathogens, including the sequence information of the genomes, distribution of the sequence types, and the collection place of the strain in the world. In Analysis page, users can upload a FASTA sequence files to analysis the genome sequence , including species identification , SNP Calling , Plot the Minimum spanning tree, phylogenetic tree analysis, finding kinship between genomes and trace the origin of pathogenic bacteria, and send a full report after the analysis task is complete.